Example Experiment: Characterizing ERK Activation in Response to FGF Treatment in NIH-3T3 Cells
Background
Accurate quantification of protein expression and/or post-translational modifications is important for advancing both basic and translational research. Given the flexibility, reproducibility, and higher throughput of the In‑Cell Western™ Assay, it offers a convenient alternative to Western blotting and is a powerful platform for meaningful in situ analyses. The In-Cell Western microplate format can be used to analyze:
- Protein phosphorylation and signaling (1 - 3)
- Off-target effects of drugs on signaling pathways (4)
- Timing and kinetics of signaling events (5, 6)
- Quantification of viral load (7 - 11)
- Genotoxicity assays (12, 13)
- Cell proliferation and apoptosis assays (14)
- Bacterial-induced epithelial signaling (15)
- Glycoprotein analysis (16, 17)
- Library screening (18 - 20)
- Screening of monoclonal antibody clones (21)
Introduction
The fibroblast growth factor (FGF) regulates numerous processes in both developing and adult tissues such as, proliferation, differentiation, migration, and cell survival by binding to the fibroblast growth factor receptor (FGFR) family, thereby triggering a signal transduction cascade that initiates a variety of biochemical and molecular changes, including the phosphorylation and activation of downstream kinases: mitogen-activated protein kinase kinase (MEK) and extracellular signal–regulated kinases (ERK) (22). The ability of the FGF pathway to direct multiple programs necessary for tissue development and regeneration, suggests that the perturbation of this pathway can promote malignant behavior in cells (23). Consequently, exploring the complex FGFR signaling pathways can provide critical knowledge in understanding both its role in both normal and pathological processes.
Experimental Design
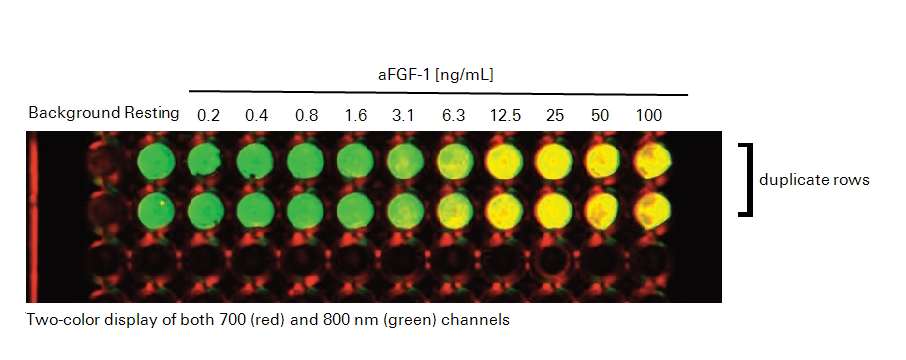
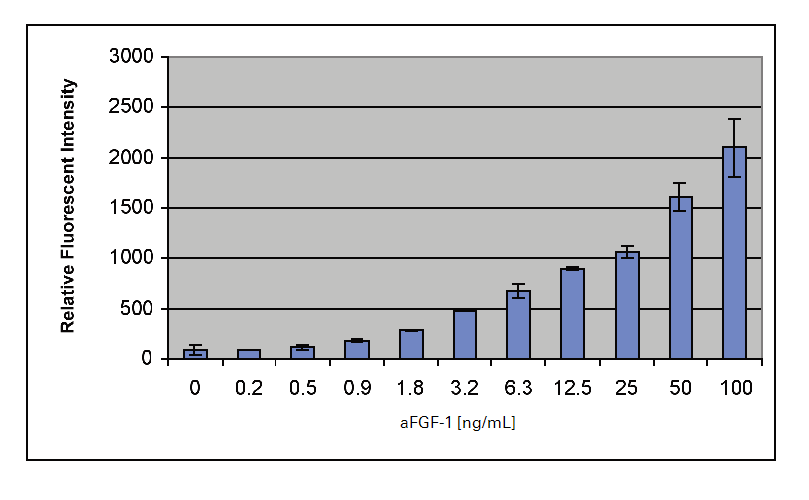
In the following In‑Cell Western™ Assay example, we present a protocol for monitoring ERK phosphorylation in response to stimulation of the FGFR pathway by the acidic fibroblast growth factor 1, αFGF-1. Changes in both phospho- and total- ERK protein levels are detected and characterized in NIH-3T3 cells treated with a serial dilution of αFGF-1. This In-Cell Western format allows for the assessment of the ERK activation, a kinase downstream in the FGFR pathway. The appropriate experimental conditions identified in this experimental protocol were used to evaluate the ability of a MEK1/2 inhibitor to modulate the FGFR pathway.
Required Reagents
LI‑COR Reagents
- IRDye® 800CW Goat anti-Rabbit Secondary Antibody (LI-COR P/N 926-32211)
- CellTag™ 700 Stain (LI-COR P/N 926-41090)
- Intercept® (PBS) Blocking Buffer (licor.com/intercept)
Additional Reagents
- 1X PBS wash buffer
- NIH-3T3 cells (ATCC, P/N CRL-1658)
- Tissue culture reagents (serum, DMEM, trypsin, 1X PBS)
- Poly-D-Lysine black-sided 96-well or 384-well microplates with clear well bottoms (Plate Handling)
Heparin (Millipore® Sigma, P/N 375095)
- Acidic Fibroblast Growth Factor (Upstate Group Inc., P/N 01-116)
- Primary antibodies
- 20% Tween® 20
- 37% formaldehyde
- 10% Triton® X-100
Plate Handling
Be extremely cautious and delicate in handling plates and pipetting to avoid detaching the cells.
NIH3T3 cells do not adhere strongly to TC-treated plates, resulting in the need for Poly-D-Lysine-coated plates in this assay. However, even with lysine-coated plates, the adherence of cells remains relatively weak compared with other cell lines.
Prepare Cells
Allow NIH3T3 cell growth in a T75 flask using standard tissue culture procedures until ~80% confluency is achieved (~1.5 x 107 cells; DMEM, 10% FBS; Gibco®).
Remove growth media and wash cells with sterile 1X PBS (room temperature (RT)).
Add trypsin and incubate 3-5 minutes at 37 °C to displace cells.
Neutralize displaced cells with culture media and pellet by centrifugation (500 x g).
Remove supernatant and disrupt the cell pellet manually by hand-tapping the collection tube.
To maintain cell integrity, do not pipet or vortex during pellet disruption.
Resuspend cells in 20 mL of complete media and count cells using a hemocytometer.
Reconstitute and dilute cells with complete media to a concentration of 75,000 cells/mL.
Manually mix the cell suspension thoroughly.
Under sterile conditions, dispense 200 µL of the cell suspension per well into a microplate (15,000 cells plated per well).
Incubate cells at 37 °C and monitor cell density until 70% confluency is achieved. This should take approximately about 24 hours.
70% confluency is very important. 90-100% confluent cells have a higher likelihood of detachment during washing.
Treat Cells
- Warm serum-free media (DMEM; Gibco®) to 37 °C.
- Remove media and inhibitor from plate wells by aspiration or manual displacement.
- Add either serum-free media for resting cells (mock) or serum-free media with serial concentrations of aFGF-1 ranging from 0.2 to 100 ng/mL, combined with 10 µg/mL heparin for activated cells. Use 100 µL of resting or activation media per well.
- Incubate at 37 °C for 7.5 minutes.
Fix and Permeabilize Cells
Fix Cells
Prepare fresh Fixing Solution as follows:
1X PBS 45 mL 37% Formaldehyde 5 mL 3.7% Formaldehyde 50 mL When incubation period is complete, carefully remove activation media manually or by aspiration to avoid detaching the cells.
Using a multi-channel pipettor, add 150 µL of fresh Fixing Solution (RT). Add the solution by pipetting down the sides of the wells carefully to avoid detaching the cells from the well bottom.
Allow incubation on bench top for 20 minutes at RT with no shaking.
Permeabilize Cells
Prepare Triton® Washing Solution as follows:
1X PBS 495 mL 10% Triton X-100 5 mL 1X PBS + 0.1% Triton X-100 500 mL Remove Fixing Solution to an appropriate waste container (contains formaldehyde).
Using a multi-channel pipettor, add 200 µL Triton Washing Solution (RT). Add the solution down the sides of the wells carefully to avoid detaching the cells.
Allow plate to shake on a rotator for 5 minutes at RT.
Repeat washing steps 4 more times, removing wash manually each time.
Do not allow cells to become dry during washing. Immediately add the next wash after manual disposal.
Block Cells
- Using a multi-channel pipettor, add 150 µL of Intercept® Blocking Buffer to each well. Add the solution by pipetting down the sides of the wells carefully to avoid detaching the cells.
- Allow blocking for 1.5 hours at RT with moderate shaking on a plate shaker.
Primary Antibodies
Dilute Primary Antibodies
Add the two primary antibodies in Intercept Blocking Buffer. Combine the following solutions for phospho-ERK target analysis:
Phospho-ERK (Rabbit; 1:100 dilution in the combined solution; Cell Signaling Technology P/N 9101)
Total ERK2 (Mouse; 1:100 dilution in the combined solution; Santa Cruz Biotechnology P/N SC-1647)
Mix the primary antibody solution thoroughly before adding to wells.
Incubate with Primary Antibodies
- Remove the blocking buffer from the wells and add 50 µL of the desired primary antibody or antibodies in Intercept Blocking Buffer to cover the bottom of each well.
- Make sure to include background wells without primary antibody to serve as a source for background well intensity. Only add 50 µL of Intercept Blocking Buffer to background wells.
- Incubate with primary antibody overnight with gentle shaking at RT.
Wash
Prepare Tween® Washing Solution as follows:
1X PBS 995 mL 20% Tween 20 5 mL 1X PBS + 0.1% Tween 20 1000 mL Remove primary antibody solution.
Using a multi-channel pipettor, add 200 µL Tween Washing Solution (RT). Add solution down the sides of the wells carefully to avoid detaching the cells from the well bottom.
Allow wash to shake on plate shaker for 5 minutes at RT.
Repeat washing steps 4 more times.
Secondary Antibodies
Dilute Secondary Antibodies
Dilute the fluorescently-labeled secondary antibodies in Intercept® Blocking Buffer as specified below. To lower background, add Tween® 20 to the diluted antibody to a final concentration of 0.2%. Recommended dilution range is 1:200 – 1:1,200.
Goat anti-Rabbit IRDye® 680RD (1:800 dilution in the combined solution)
Goat anti-Mouse IRDye 800CW (1:800 dilution in the combined solution)
Minimize exposure of the antibody vials to light.
Mix the antibody solutions and add 50 µL of the secondary antibody solution to each well.
Incubate with Secondary Antibodies
- Incubate for 60 minutes with gentle shaking at RT. Protect plate from light during incubation.
Wash
- Remove secondary antibody solution.
- Using a multi-channel pipettor, add 200 µL of Tween Washing Solution at RT (Wash). Add solution down the sides of the wells carefully to avoid detaching the cells from the well bottom.
- Allow wash to shake on a plate shaker for 5 minutes at RT.
- Repeat washing steps 4 more times. Protect plate from light during washing.
Image
- After final wash, remove wash solution completely from wells. Turn the plate upside down and tap or blot gently on paper towels to remove traces of wash buffer. For best results, scan plate immediately; plates may also be stored at 4 °C for several weeks (sealed and protected from light).
- Before plate scanning, clean the bottom plate surface and the Odyssey® Imager scanning bed (if applicable) with moist, lint-free tissue to avoid any obstructions during scanning.
- Scan plate with detection in both 700 and 800 nm channels.
Suggested Scan Settings
All settings may require adjustment for optimal data quality. Higher resolutions or scan qualities can be used, but the scan time will increase.
| Instrument | Resolution | Scan Quality | Intensity Setting (700 nm) | Intensity Setting (800 nm) |
| Odyssey Classic | 169 µM | lowest | 5 | 5 |
| Odyssey DLx | 169 µM | lowest | Auto Mode | Auto Mode |
| Odyssey Sa | 200 µM | lowest | 7 | 7 |
| Aerius™ Imager | 200 µM | lowest | 7 | 7 |
Experimental Results
References
1. Chen, H., Kovar, J., Sissons, S., Cox, K., Matter, W., Chadwell, F., Luan, P., Vlahos, C. J., Schutz-Geschwender, A., and Olive, D. M. (2005) A cell-based immunocytochemical assay for monitoring kinase signaling pathways and drug efficacy. Analytical biochemistry 338, 136-142
2. Aguilar, H. N., Zielnik, B., Tracey, C. N., and Mitchell, B. F. (2010) Quantification of rapid Myosin regulatory light chain phosphorylation using high-throughput in-cell Western assays: comparison to Western immunoblots. PLoS One 5, e9965
3. Wong, S. K. (2004) A 384-well cell-based phospho-ERK assay for dopamine D2 and D3 receptors. Analytical biochemistry 333, 265-272
4. Kumar, N., Afeyan, R., Kim, H. D., and Lauffenburger, D. A. (2008) Multipathway model enables prediction of kinase inhibitor cross-talk effects on migration of Her2-overexpressing mammary epithelial cells. Mol Pharmacol 73, 1668-1678
5. Hannoush, R. N. (2008) Kinetics of Wnt-driven beta-catenin stabilization revealed by quantitative and temporal imaging. PLoS One 3, e3498
6. Chen, W. W., Schoeberl, B., Jasper, P. J., Niepel, M., Nielsen, U. B., Lauffenburger, D. A., and Sorger, P. K. (2009) Input-output behavior of ErbB signaling pathways as revealed by a mass action model trained against dynamic data. Mol Syst Biol 5, 239
7. Counihan, N. A., Daniel, L. M., Chojnacki, J., and Anderson, D. A. (2006) Infrared fluorescent immunofocus assay (IR-FIFA) for the quantitation of non-cytopathic and minimally cytopathic viruses. J Virol Methods 133, 62-69
8. Lin, Y. C., Li, J., Irwin, C. R., Jenkins, H., DeLange, L., and Evans, D. H. (2008) Vaccinia virus DNA ligase recruits cellular topoisomerase II to sites of viral replication and assembly. J Virol 82, 5922-5932
9. Weldon, S. K., Mischnick, S. L., Urlacher, T. M., and Ambroz, K. L. (2010) Quantitation of virus using laser-based scanning of near-infrared fluorophores replaces manual plate reading in a virus titration assay. J Virol Methods 168, 57-62
10. Lopez, T., Silva-Ayala, D., Lopez, S., and Arias, C. F. (2012) Methods suitable for high-throughput screening of siRNAs and other chemical compounds with the potential to inhibit rotavirus replication. J Virol Methods 179, 242-249
11. Wan, Y., Zhou, Z., Yang, Y., Wang, J., and Hung, T. (2010) Application of an In-Cell Western assay for measurement of influenza A virus replication. J Virol Methods 169, 359-364
12. Jamin, E. L., Riu, A., Douki, T., Debrauwer, L., Cravedi, J. P., Zalko, D., and Audebert, M. (2013) Combined genotoxic effects of a polycyclic aromatic hydrocarbon (B(a)P) and an heterocyclic amine (PhIP) in relation to colorectal carcinogenesis. PLoS One 8, e58591
13. Khoury, L., Zalko, D., and Audebert, M. (2013) Validation of high-throughput genotoxicity assay screening using gammaH2AX in-cell western assay on HepG2 cells. Environ Mol Mutagen 54, 737-746
14. Godin-Heymann, N., Ulkus, L., Brannigan, B. W., McDermott, U., Lamb, J., Maheswaran, S., Settleman, J., and Haber, D. A. (2008) The T790M "gatekeeper" mutation in EGFR mediates resistance to low concentrations of an irreversible EGFR inhibitor. Mol Cancer Ther 7, 874-879
15. Du, Y., Danjo, K., Robinson, P. A., and Crabtree, J. E. (2007) In-Cell Western analysis of Helicobacter pylori-induced phosphorylation of extracellular-signal related kinase via the transactivation of the epidermal growth factor receptor. Microbes Infect 9, 838-846
16. McInerney, M. P., Pan, Y., Short, J. L., and Nicolazzo, J. A. (2017) Development and Validation of an In-Cell Western for Quantifying P-Glycoprotein Expression in Human Brain Microvascular Endothelial (hCMEC/D3) Cells. J Pharm Sci 106, 2614-2624
17. Urlacher T, Xing K, Cheung L et al (2013) Glycoprotein applications using near-infrared detection. Poster presentation, Experimental Biology
18. Guo, K., Shelat, A. A., Guy, R. K., and Kastan, M. B. (2014) Development of a cell-based, high-throughput screening assay for ATM kinase inhibitors. J Biomol Screen 19, 538-546
19. Hoffman, G. R., Moerke, N. J., Hsia, M., Shamu, C. E., and Blenis, J. (2010) A high-throughput, cell-based screening method for siRNA and small molecule inhibitors of mTORC1 signaling using the In Cell Western technique. Assay Drug Dev Technol 8, 186-199
20. Schnaiter, S., Furst, B., Neu, J., Waczek, F., Orfi, L., Keri, G., Huber, L. A., and Wunderlich, W. (2014) Screening for MAPK modulators using an in-cell western assay. Methods Mol Biol 1120, 121-129
21. Daftarian, M. P., Vosoughi, A., and Lemmon, V. (2014) Gene-based vaccination and screening methods to develop monoclonal antibodies. Methods Mol Biol 1121, 337-346
22. Ornitz, D. M., and Itoh, N. (2015) The Fibroblast Growth Factor signaling pathway. Wiley Interdiscip Rev Dev Biol 4, 215-266
23. Babina, I. S., and Turner, N. C. (2017) Advances and challenges in targeting FGFR signalling in cancer. Nat Rev Cancer 17, 318-332